Controlled vocabularies such as the Gene Ontology enable you to compare large numbers of genes and find differences in gene content. In this exercise we will compare the gene content of two fungi.
1. Programs such as PoGO and BLAST2GO enable you to automatically predict GO terms for proteins. Unfortunately, these programs take too long to run, so we cannot use them during the class. The following files are examples of reports made by PoGO or BLAST2GO: Saccharomyces cerevisiae: yeast.txt and Schizosaccaromyces pombe: spombe.txt
Download both of these files to your computer. Double-click them to uncompress them and then open them with the TextEdit program. Notice that the data are organized into two columns. Any program can be used to create GO terms for a list of programs but the GO terms must be organized into two columns so that they can be used with GOSlimViewer.
2. We will use the GOSlimViewer program at AGBase to compare the GO annotations shown in step 1 to a GO slim file. On the GOSLimViewer website, select one of the GO report files and then select the yeast GO Slim set. Click the search button.
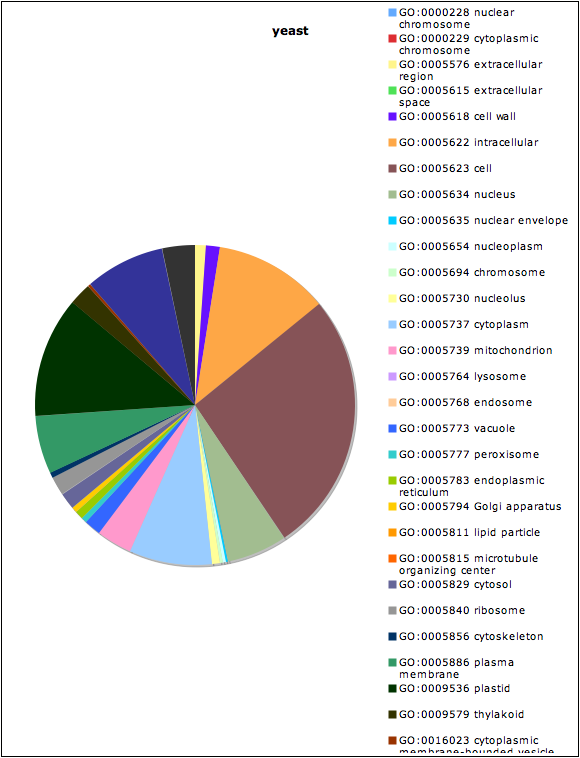
3. When the analysis has finished, the web site will show you three links. Select one of the links to see a table with GO Slim terms and a number thar represents the number of genes with each GO Slim term.
4. Optional: Copy and paste the table in step 3 into an Excel spreadsheet. You can use excel to make various types of figures using this data source, such as a pie chart