Plant diseases are one of the main limiting factors to agricultural production world-wide and our ability to feed the world’s growing population will depend on finding better methods to control disease. Our research program is focused on the comparative genomics of filamentous fungi, with the goal of understanding the evolution of pathogenicity in plant pathogenic fungi. Plant pathogenicity is a highly variable trait and at most taxonomic levels within the fungi we can find both pathogens and non-pathogens. By studying how fungi have evolved to infect plants and cause diseases, we hope to identify new aspects of the disease process that can be leveraged for developing improved control methods.
We study various aspects of plant pathology, genetics and plant-fungus interactions, including hemibiotrophic pathogenesis, utilizing the model Colletotrichum graminicola-maize. More information about Colletotrichum graminicola can be found on our maize anthracnose page. Our main lines of research are:
Effectoromics
In recent years we have begun to recognize the important role that effectors have in mediating the apparently complex interchange of signals between plant and pathogen. The identification of genes encoding effector proteins has become one of plant pathology’s hot topics, as evidenced by the number of manuscripts on the subject that have been published in recent years. Previous studies have shown that in some plant pathogens, effector proteins evolve at accelerated rates compared to other genes in the genome. We are using comparative genomics to identify gene families with the signatures of accelerated evolution with the goal of finding novel effectors in C. graminicola. In the laboratory, we are using molecular and cellular biological methods to study the putative effectors that we identify through comparative genomics.
Comparative and Evolutionary Genomics
We study the evolution of fungal genomes, with the goal of better understanding how pathogenicity has evolved in fungi. Some of the topics that we are currently working on include:
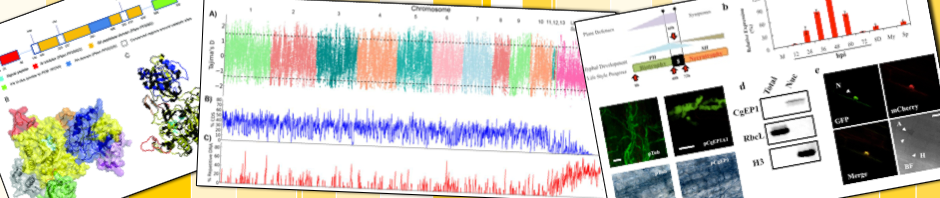
Natural selection acting on fungal genomes. Natural selection is expected to leave contrasting imprints on DNA, offering the opportunity to identify functionally important genomic regions and interpret genetic variations shaping organismal phenotypes. For pathogens, identifying the mode of evolution within the genome can aide in the pursuit of effective strategies to control diseases.
Molecular Mimicry. Proteins produced by pathogens can mimic proteins of the host, and thereby bind to host receptors or otherwise cause changes in the host metabolism. Molecular mimicry has been recognized for some time. Oldstone (1998) defines molecular mimicry as, “… similar structures shared by molecules from dissimilar genes or by their protein products. Either the molecules’ linear amino acid sequences or their conformational fits may be shared, even though their origins are as separate as, for example, a virus and a normal host–self determinant.” The extent to which fungal proteins mimic those of plants is unknown. In one line of research, we plan to identify putative protein mimics encoded by genes in the C. graminicola genome.
Horizontal Gene Transfer In Fungi. In another line of research, we are identifying the extent to which horizontal gene transfer has impacted the gene content of Colletotrichum spp. Horizontal gene transfer (HGT) referrers to the transfer of genetic material between species using mechanisms other than inheritance from ancestors. HGT is relatively rare in higher eukaryotes, but is more common in microbes such as bacteria and archaea. The horizontal transfer of DNA between bacteria and archaea is now a well established phenomenon. In fungi, HGT has been used to explain the similarity of secondary metabolite gene clusters among species. Horizontally transferred genes may allow the pathogen to mimic either the plant’s own proteins, or those of other organisms. However, horizontally transferred genes may serve other functions as well. They may enable the pathogen to synthesize new toxins, or allow it to metabolize new substrates. The identification of HGT in Colletotrichum spp. will provide important insight into genome evolution as well as help to understand the evolution of traits that play a role in plant-pathogen interactions. In this line of research, we are using computational methods to identify the horizontally transferred genes in the genomes of Colletotrichum spp.
Current Research Grants
- Characterization and comparative genomic analysis of pathogenicity in the maize anthracnose fungus Colletotrichum graminicola. Junta de Castilla y León. 1/2013 to 9/2016.
- GEN2PHEN: From genotype to phenotype: Integrated population genomic and molecular genetic studies of anthranose causing fungi. Ministerio de Economía y Competitividad (AGL2015-66362-R) 01/2016 – 12/2018